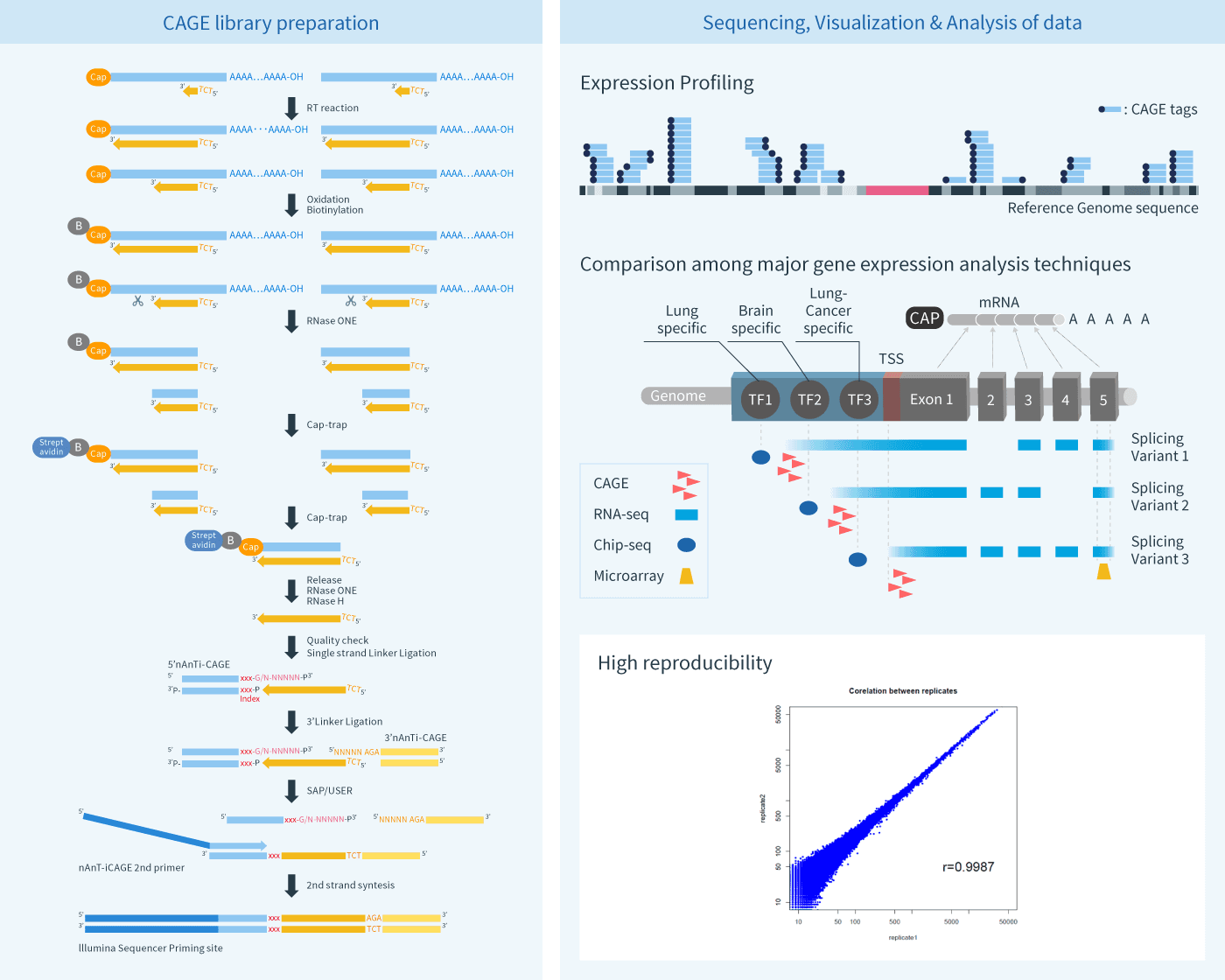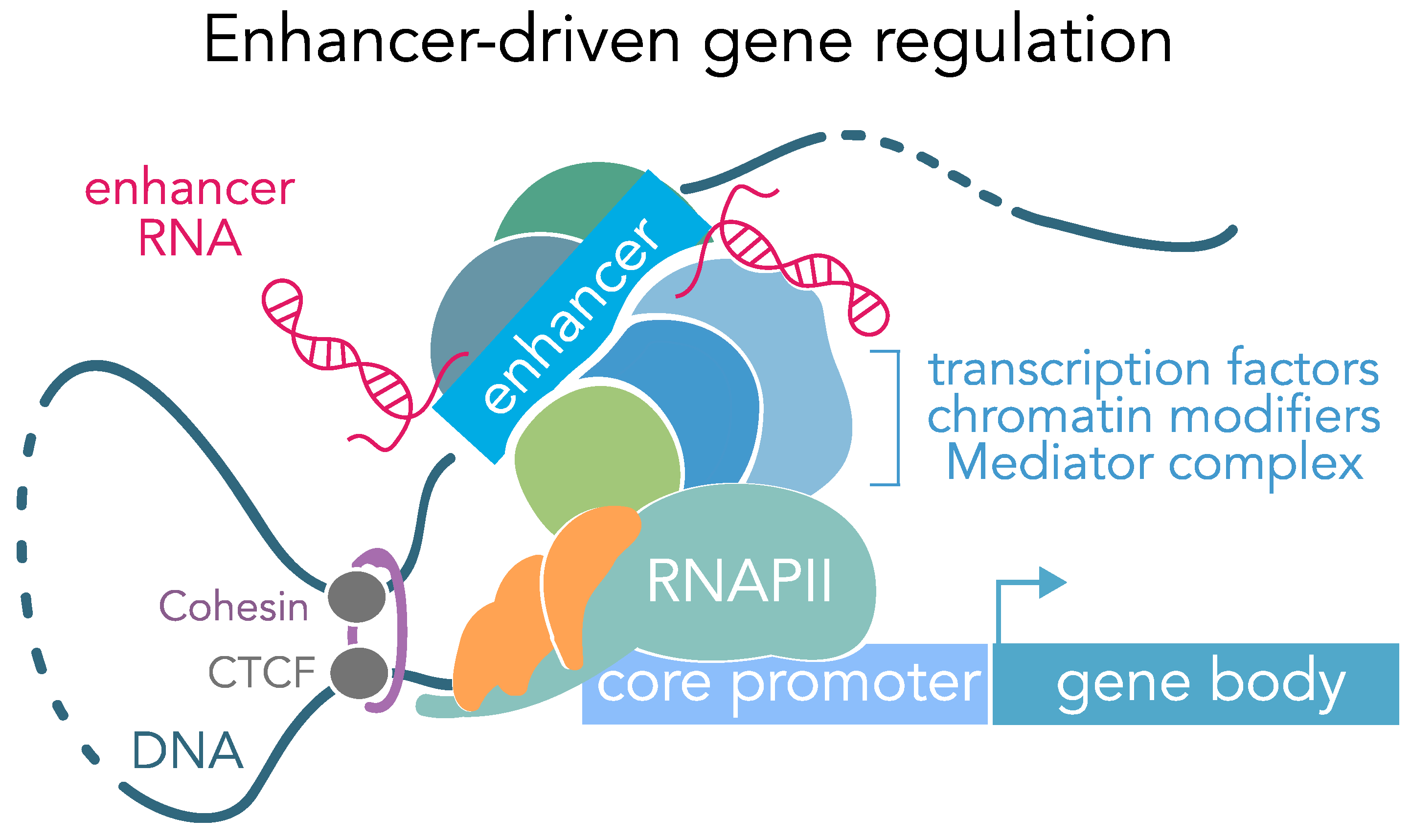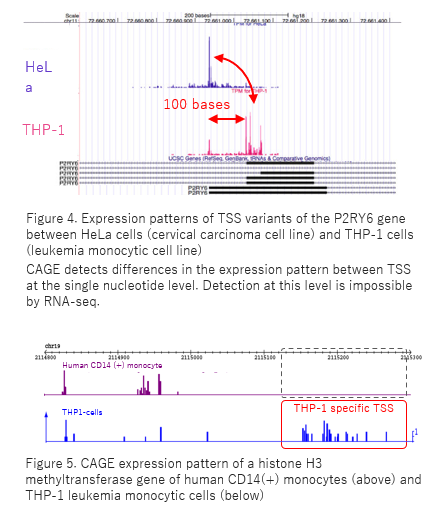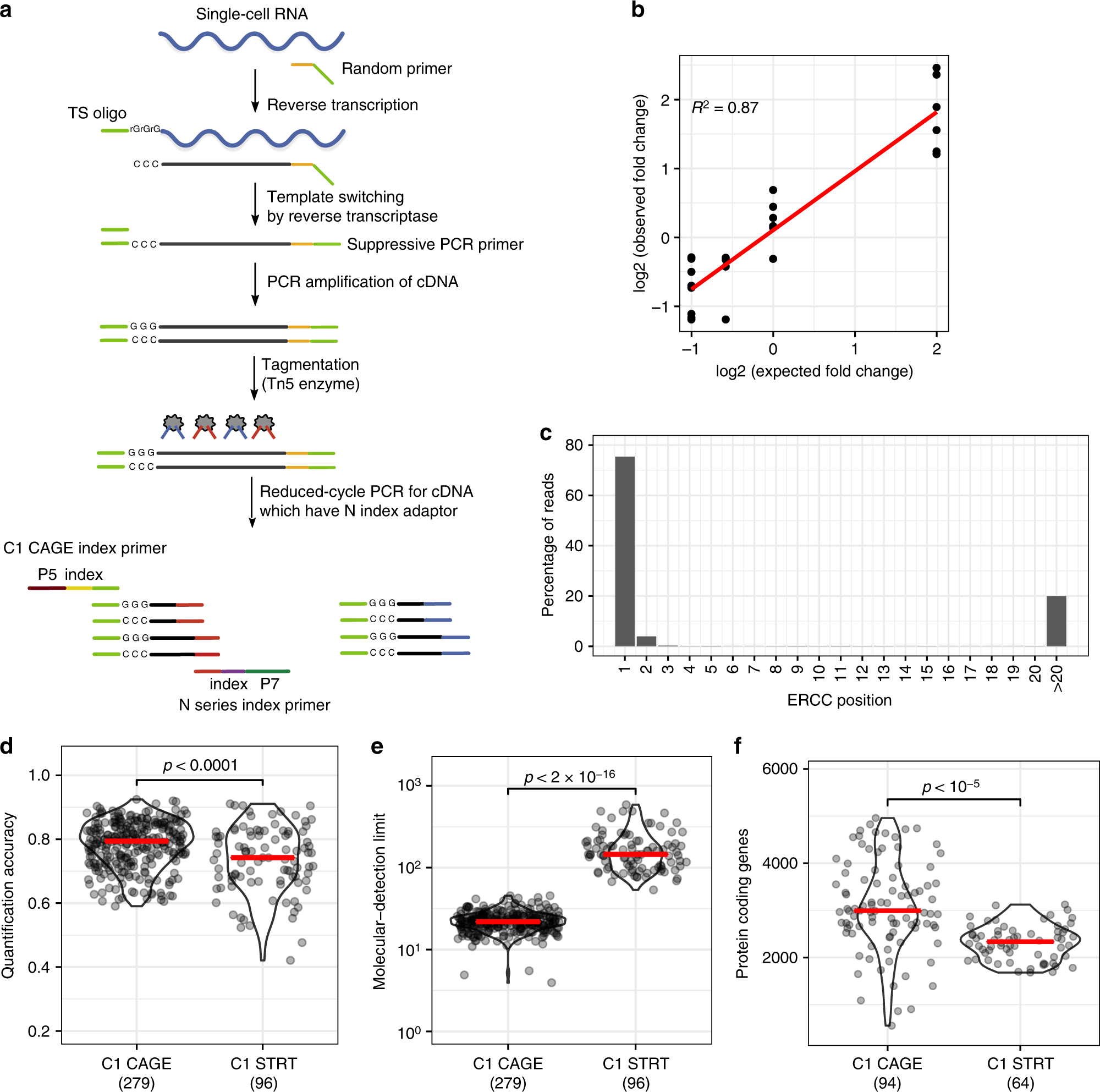
C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution | Nature Communications

A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super- enhancer Activities in Cancer - ScienceDirect

A High-Resolution Map of Human Enhancer RNA Loci Characterizes Super- enhancer Activities in Cancer - ScienceDirect

Landscape of Enhancer-Enhancer Cooperative Regulation during Human Cardiac Commitment: Molecular Therapy - Nucleic Acids
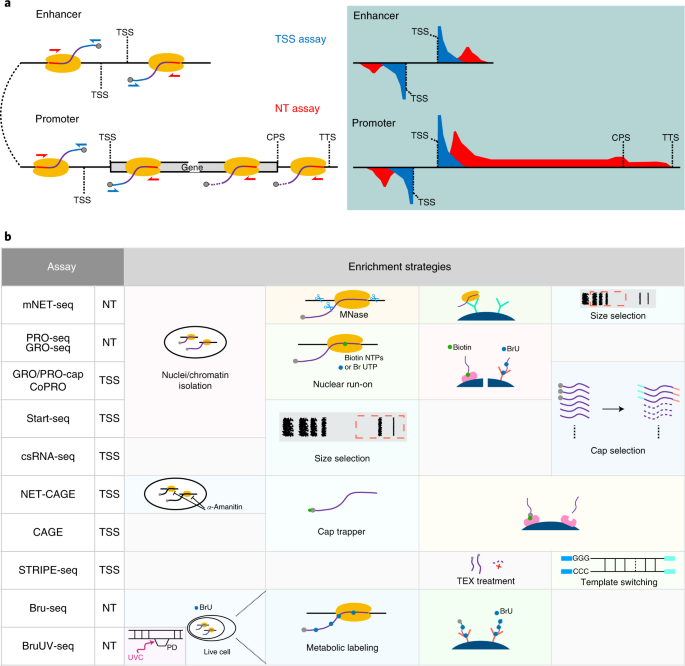
A comparison of experimental assays and analytical methods for genome-wide identification of active enhancers | Nature Biotechnology

Identification of P300 dense super-enhancers. a Ranked distribution... | Download Scientific Diagram
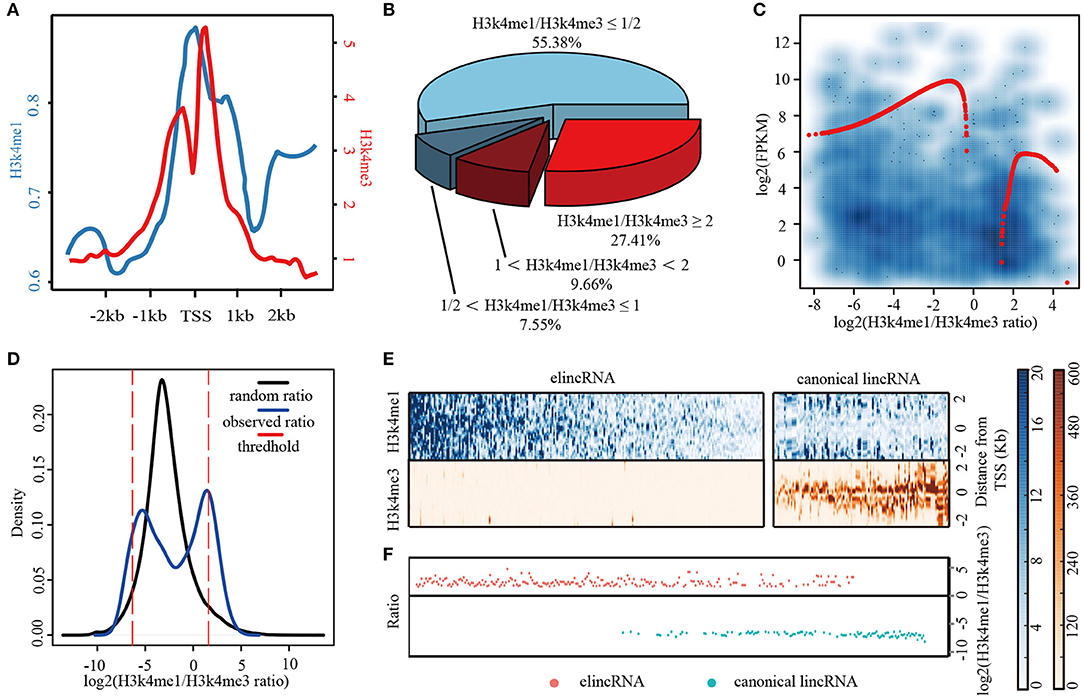
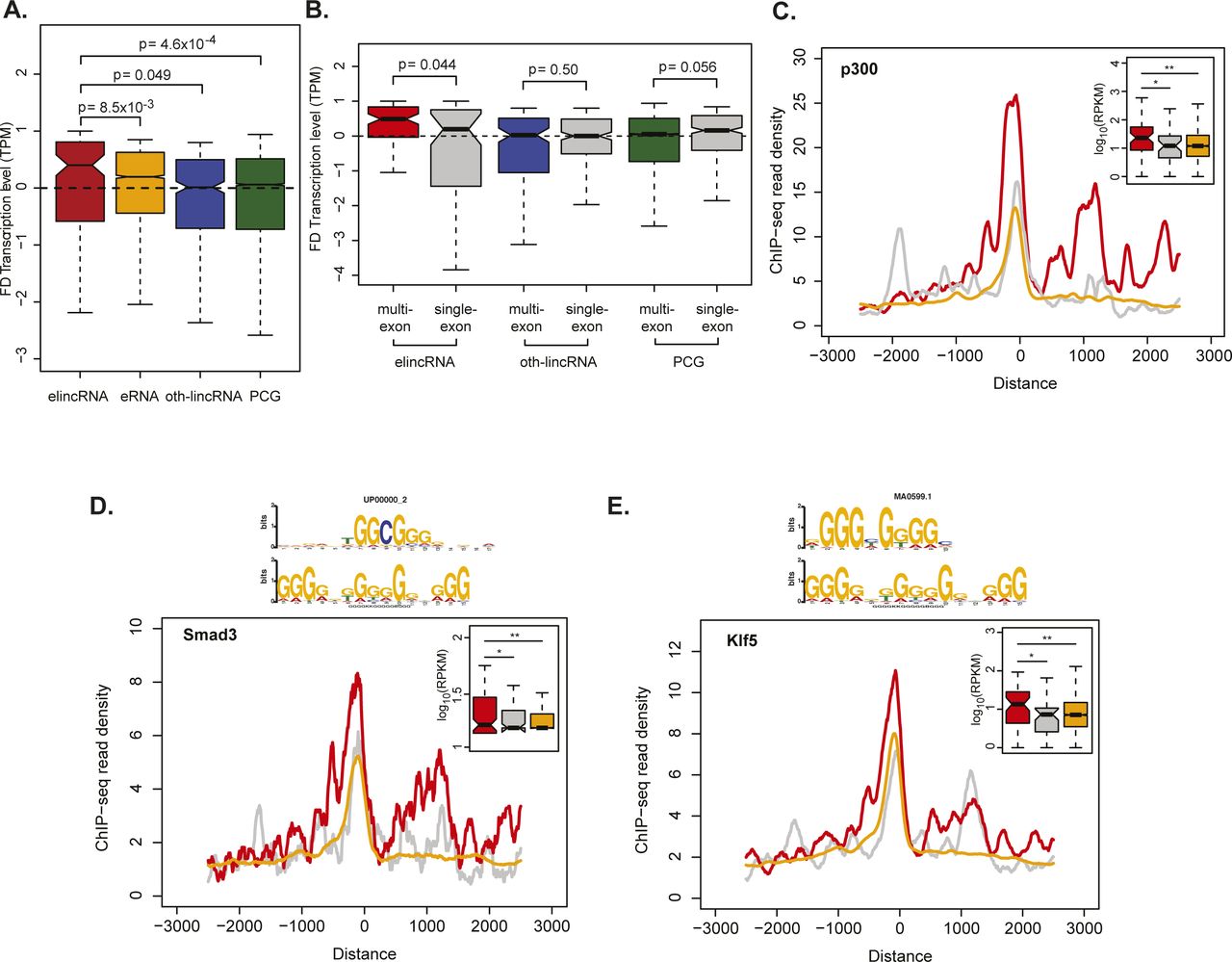
Frontiers | A Novel Approach to Identify Enhancer lincRNAs by Integrating Genome, Epigenome, and Regulatome | Bioengineering and Biotechnology

Genome-wide enhancer annotations differ significantly in genomic distribution, evolution, and function | BMC Genomics | Full Text

Enhancer and super‐enhancer: Positive regulators in gene transcription - Peng - 2018 - Animal Models and Experimental Medicine - Wiley Online Library

Enhancer and super‐enhancer: Positive regulators in gene transcription - Peng - 2018 - Animal Models and Experimental Medicine - Wiley Online Library

Features of eSTARR-seq enhancers a, Scatterplot of activity vs GRO-cap... | Download Scientific Diagram

Genome-wide identification of ESR1 enhancer transcripts in MCF-7 cells... | Download Scientific Diagram

Frontiers | Identification and Analysis of p53-Regulated Enhancers in Hepatic Carcinoma | Bioengineering and Biotechnology

NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements | Nature Genetics